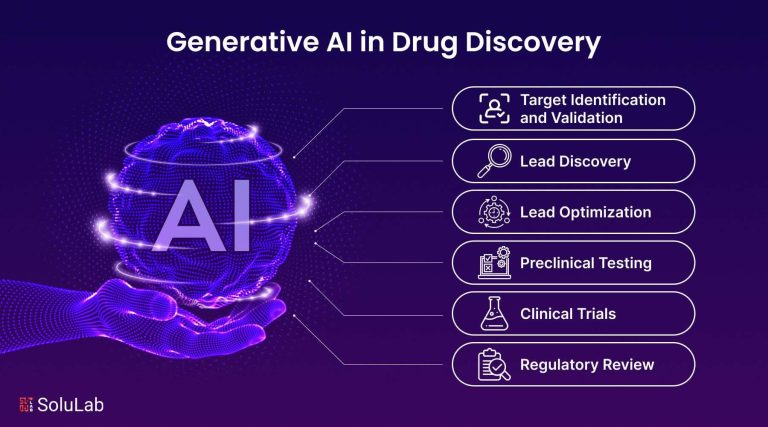
Artificial intelligence is accelerating the front end of drug development, compressing tasks that once took years into months and pushing experimental compounds into the clinic faster than traditional methods. After years of pilots, algorithm-driven discovery is moving into pharma’s mainstream, as global drugmakers strike multi‑year deals with AI platforms, hire in‑house model teams, and retool labs for data-centric R&D.
What’s changing is not just speed but scope: generative models design novel molecules, structure-prediction tools expand the target universe, and closed-loop systems pair machine learning with automated synthesis and testing. The stakes are high. With patent cliffs approaching and R&D costs climbing, companies are betting that AI can raise success rates as well as shorten timelines. Regulators are sharpening guidance on AI/ML use, investors are flooding the sector, and early AI-derived candidates are entering human studies-setting up a pivotal test of whether the technology can deliver safer, more effective medicines at scale.
Table of Contents
- AI compresses early discovery timelines as foundation models pinpoint targets and design novel molecules
- Pharma shifts investment to data pipelines and model operations to turn pilots into production
- Regulators spotlight transparency with validation benchmarks, reproducible workflows and auditable model traces
- Action plan Build cross functional AI squads, institute model risk governance and join data sharing consortia
- Key Takeaways
AI compresses early discovery timelines as foundation models pinpoint targets and design novel molecules
Pharma teams are shrinking the hypothesis-to-hit window as large, multimodal foundation models mine literature, omics, structures, and real-world data to surface causal biology and tractable targets. By fusing language, graph, and protein models, platforms now score target druggability, predict binding sites, and propose lead-like scaffolds in the same loop, enabling rapid triage and design cycles that previously took months. Early deployments report tighter prioritization, fewer dead ends, and faster transition to in vitro validation, with de novo design and ADMET-aware optimization arriving earlier in the funnel.
- Target identification: cross-evidence reasoning from genetics, pathway maps, and patient stratification.
- Pocket prediction: structure and dynamics inference for harder proteins, including cryptic sites.
- Molecule generation: generative models craft synthetically tractable chemotypes with property constraints.
- Multi-objective optimization: potency, selectivity, and safety balanced via fast surrogate models.
- Synthesis planning: AI-guided retrosynthesis aligned to available building blocks.
Operationally, discovery groups are stitching together data fabrics, cloud-scale training, and closed-loop DMTA with robotic labs, enabling dozens of design-test iterations per week and earlier kill-or-commit decisions. Governance is moving in lockstep: model provenance, uncertainty estimates, and assay concordance are becoming standard, as are cross-functional reviews to de-risk bias and ensure reproducibility ahead of IND-enabling studies.
- Key enablers: multimodal data unification, active learning, and synthesis-aware generative models.
- Quality controls: calibration against held-out assays, orthogonal validation, and audit-ready model cards.
- Impact signals: fewer cycles to a qualified hit, higher validated target rate, and reduced attrition pre-IND.
- Scale-up: APIs into ELNs/LIMS, automated reporting, and cost-aware compute scheduling.
Pharma shifts investment to data pipelines and model operations to turn pilots into production
Major drugmakers are redirecting budgets from flashy pilots to production-grade data infrastructure, seeking reliability over novelty. Executives describe a pivot toward standardized data pipelines that can withstand audits, span discovery-to-development workflows, and integrate lab instruments with clinical and real‑world data. The goal: remove bottlenecks that stalled early AI wins, enforce FAIR-by-design practices, and make models retrainable, traceable, and compliant across global sites. Procurement teams are consolidating tools, while IT and R&D align on KPIs such as cycle-time reduction from hit identification to candidate selection and model reusability across programs.
- Data fabrics and lakehouse architectures: harmonizing chemistry, omics, imaging, and EHR streams with governed metadata
- ELT/ETL orchestration: cloud-native pipelines with lineage, versioning, and automated quality gates
- Feature stores and vector indexes: reusable representations for generative and predictive models
- Privacy-by-design: tokenization, federated learning, and synthesis to enable multi-party analyses
- GxP-readiness: audit trails, role-based controls, and validated connectors to lab and clinical systems
Alongside data modernization, companies are institutionalizing model operations (MLOps) to move algorithms out of notebooks and into validated environments. New operating models pair data engineers with pharmacometricians and biologists, enforcing continuous validation, policy-as-code, and explainability in decision points that affect trial design and portfolio choices. Real‑time monitoring flags drift as assays evolve, while registries track lineage from training data to deployment. In labs, closed‑loop workflows link in silico screens to robotics, turning predictions into experiments at scale.
- Model registries and CI/CD for ML: standardized promotion paths from sandbox to production
- Risk controls: bias assessments, performance SLAs, and change-management playbooks
- Monitoring & alerting: data drift, concept drift, and stability checks tied to retraining triggers
- Decision traceability: documentation that supports inspections and cross-study reproducibility
- Business impact: faster lead optimization, reduced assay reruns, and earlier go/no‑go clarity in R&D
Regulators spotlight transparency with validation benchmarks, reproducible workflows and auditable model traces
Regulatory agencies across the U.S., EU and U.K. are converging on clearer expectations for AI in pharma R&D, signaling the end of opaque, black‑box discovery workflows. Sponsors report that filings increasingly hinge on defensible, cross‑site performance evidence and explicit governance of data lineage. Submissions are expected to pair headline metrics with context, including external validation and stress testing. Requirements frequently cited by reviewers include:
- Benchmark rigor: pre‑defined baselines, multi‑center hold‑out datasets, and shift/robustness analyses under realistic lab and clinical variability.
- Fairness and uncertainty: subgroup performance, calibration, and exposure of confidence intervals for decision thresholds that affect downstream experiments.
- Documentation: traceable model cards and data cards that specify provenance, exclusions, and known failure modes.
Execution standards are tightening as well, with inspectors prioritizing reproducibility and traceability from hypothesis generation to candidate nomination. Reviewers now look for deterministic, containerized pipelines and full auditability of model behavior over time. Sponsors preparing for inspections are building operational controls that make AI‑enabled discovery repeatable, explainable and compliant with GxP‑aligned practices:
- Reproducible workflows: version‑locked code, datasets and hyperparameters; seeded runs; container images; and signed SBOMs for models and dependencies.
- Auditable traces: end‑to‑end lineage of inputs and outputs, feature attribution records, prompt/output logs for generative tools, and cryptographic provenance of intermediate artifacts.
- Change control and surveillance: pre‑specified update plans, comparability protocols, and continuous post‑deployment monitoring with alerting when performance drifts or subgroups degrade.
Action plan Build cross functional AI squads, institute model risk governance and join data sharing consortia
Pharma leaders are operationalising AI by moving from siloed pilots to product-centric squads that pair scientists with technologists under clear accountability. Standing teams blend medicinal chemistry, biology, clinical, data engineering and MLOps to ship validated models into lab and trial workflows, with product owners measured on R&D outcomes-not demo counts. Early wins hinge on disciplined delivery: short sprints, shared backlogs with therapeutic area heads, and infrastructure that makes model reuse safer and faster across programs.
- Form mission-driven squads: computational chemists, translational scientists, clinicians, data engineers, ML researchers, MLOps, QA/RA, and security under a single product owner.
- Adopt a common toolchain: feature stores, experiment tracking, model registry, and CI/CD aligned to GxP contexts; secure sandboxes for vendor models.
- Sprint on measurable outcomes: reduce design-make-test-analyse cycle time, increase hit rate, cut FTE hours per candidate, and improve protocol feasibility scores.
- Embed change management: SOPs, training, and user feedback loops to drive lab and clinical adoption, with success tied to budget and incentives.
As models influence go/no-go decisions, robust guardrails become a board-level imperative, and shared data assets emerge as a competitive accelerant. Companies are instituting model risk governance-covering inventory, validation, bias testing, and drift monitoring-while joining precompetitive consortia to access diverse, FAIR-aligned datasets via privacy-preserving methods. The result: faster, safer deployment and broader external signal without compromising IP or patient privacy.
- Stand up model risk governance: model catalog with “model cards,” independent validation, GxP alignment, audit trails (e.g., 21 CFR Part 11), and human-in-the-loop checkpoints for high-impact use cases.
- Monitor in production: performance and data drift alerts, lineage tracking, controlled retraining, and red-teaming for robustness and security.
- Join and shape consortia: participate in precompetitive initiatives (e.g., target/disease knowledge graphs, real-world data networks), adopting FAIR and standards such as CDISC, OMOP, and FHIR.
- Protect privacy by design: data use agreements, governance charters, and techniques like federated learning, differential privacy, and secure multiparty computation to enable cross-institutional discovery.
Key Takeaways
As algorithms move from compound triage to trial design, the center of gravity in R&D is shifting. The bottlenecks are, too-from synthesis and screening to data quality, model transparency and regulatory acceptance. With regulators outlining guidance for AI-enabled submissions and companies wiring labs for closed-loop experiments, the sector is entering a proving phase.
The measure of success now extends beyond faster hit discovery. If productivity gains persist into the clinic, impact will be seen in time-to-IND, attrition rates and, ultimately, patient outcomes. AI is unlikely to replace wet science so much as recalibrate it, rewarding those who pair well-governed data with rigorous validation. For an industry under pressure on costs and timelines, the question is no longer whether AI belongs in pharma R&D, but how quickly-and safely-it can bring new medicines to patients.